pyvista.DataSetFilters.threshold#
- DataSetFilters.threshold(
- value: float | VectorLike[float] | None = None,
- scalars: str | None = None,
- invert: bool = False,
- continuous: bool = False,
- preference: Literal['point', 'cell'] = 'cell',
- all_scalars: bool = False,
- component_mode: Literal['component', 'all', 'any'] = 'all',
- component: int = 0,
- method: Literal['upper', 'lower'] = 'upper',
- progress_bar: bool = False,
Apply a vtkThreshold filter to the input dataset.
This filter will apply a vtkThreshold filter to the input dataset and return the resulting object. This extracts cells where the scalar value in each cell satisfies the threshold criterion. If
scalarsisNone, the input’s active scalars array is used.Warning
Thresholding is inherently a cell operation, even though it can use associated point data for determining whether to keep a cell. In other words, whether or not a given point is included after thresholding depends on whether that point is part of a cell that is kept after thresholding.
Please also note the default
preferencechoice for CELL data over POINT data. This is contrary to most other places in PyVista’s API where the preference typically defaults to POINT data. We chose to prefer CELL data here so that if thresholding by a named array that exists for both the POINT and CELL data, this filter will default to the CELL data array while performing the CELL-wise operation.- Parameters:
- value
float| sequence[float],optional Single value or
(min, max)to be used for the data threshold. If a sequence, then length must be 2. If no value is specified, the non-NaN data range will be used to remove any NaN values. Please reference themethodparameter for how single values are handled.- scalars
str,optional Name of scalars to threshold on. Defaults to currently active scalars.
- invertbool, default:
False Invert the threshold results. That is, cells that would have been in the output with this option off are excluded, while cells that would have been excluded from the output are included.
- continuousbool, default:
False When True, the continuous interval [minimum cell scalar, maximum cell scalar] will be used to intersect the threshold bound, rather than the set of discrete scalar values from the vertices.
- preference
str, default: ‘cell’ When
scalarsis specified, this is the preferred array type to search for in the dataset. Must be either'point'or'cell'. Throughout PyVista, the preference is typically'point'but since the threshold filter is a cell-wise operation, we prefer cell data for thresholding operations.- all_scalarsbool, default:
False If using scalars from point data, all points in a cell must satisfy the threshold when this value is
True. WhenFalse, any point of the cell with a scalar value satisfying the threshold criterion will extract the cell. Has no effect when using cell data.- component_mode{‘component’, ‘all’, ‘any’}
The method to satisfy the criteria for the threshold of multicomponent scalars. ‘component’ (default) uses only the
component. ‘all’ requires all components to meet criteria. ‘any’ is when any component satisfies the criteria.- component
int, default: 0 When using
component_mode='component', this sets which component to threshold on.- method
str, default: ‘upper’ Set the threshold method for single-values, defining which threshold bounds to use. If the
valueis a range, this parameter will be ignored, extracting data between the two values. For single values,'lower'will extract data lower than thevalue.'upper'will extract data larger than thevalue.- progress_barbool, default:
False Display a progress bar to indicate progress.
- value
- Returns:
pyvista.UnstructuredGridDataset containing geometry that meets the threshold requirements.
See also
threshold_percentThreshold a dataset by a percentage of its scalar range.
extract_values()Threshold-like filter for extracting specific values and ranges.
image_threshold()Similar method for thresholding
ImageData.select_values()Threshold-like filter for
ImageDatato keep some values and replace others.- Compare threshold filters
This example showcases this filter and other similar ones.
Examples
>>> import pyvista as pv >>> import numpy as np >>> volume = np.zeros([10, 10, 10]) >>> volume[:3] = 1 >>> vol = pv.wrap(volume) >>> threshed = vol.threshold(0.1) >>> threshed UnstructuredGrid (...) N Cells: 243 N Points: 400 X Bounds: 0.000e+00, 3.000e+00 Y Bounds: 0.000e+00, 9.000e+00 Z Bounds: 0.000e+00, 9.000e+00 N Arrays: 1
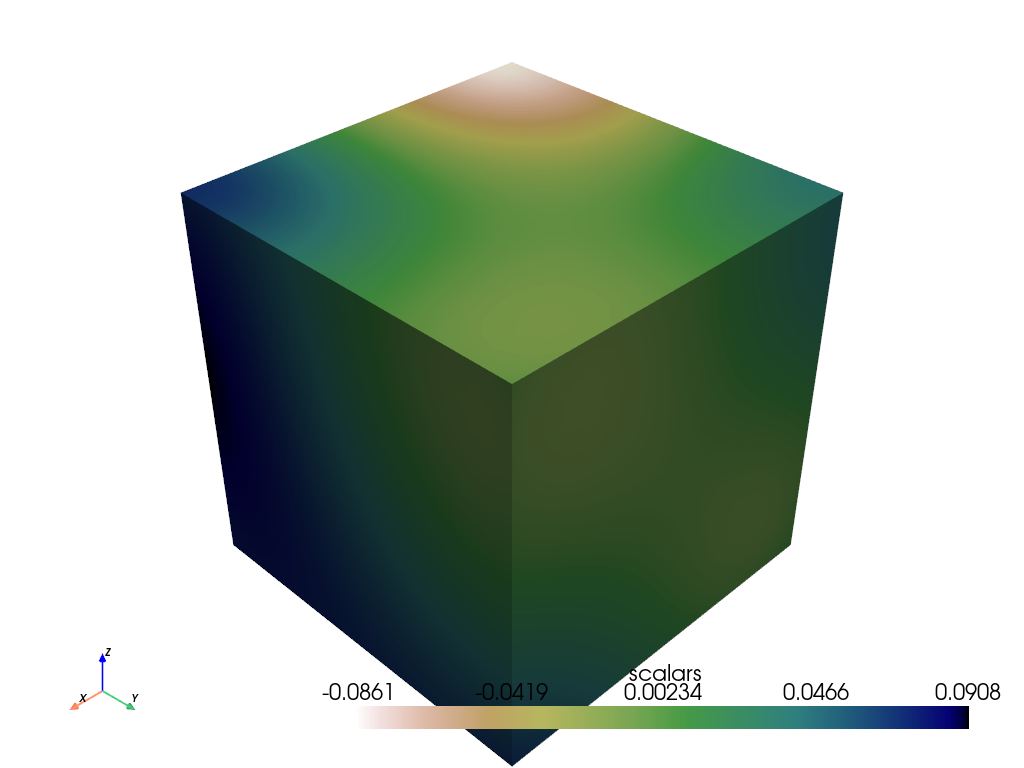
Apply the threshold filter to Perlin noise. First generate the structured grid.
>>> import pyvista as pv >>> noise = pv.perlin_noise(0.1, (1, 1, 1), (0, 0, 0)) >>> grid = pv.sample_function( ... noise, bounds=[0, 1.0, -0, 1.0, 0, 1.0], dim=(20, 20, 20) ... ) >>> grid.plot( ... cmap='gist_earth_r', ... show_scalar_bar=True, ... show_edges=False, ... )
Next, apply the threshold.
>>> import pyvista as pv >>> noise = pv.perlin_noise(0.1, (1, 1, 1), (0, 0, 0)) >>> grid = pv.sample_function( ... noise, bounds=[0, 1.0, -0, 1.0, 0, 1.0], dim=(20, 20, 20) ... ) >>> threshed = grid.threshold(value=0.02) >>> threshed.plot( ... cmap='gist_earth_r', ... show_scalar_bar=False, ... show_edges=True, ... )

See Using Common Filters and Image Data Representations for more examples using this filter.