Note
Go to the end to download the full example code.
Mesh Validation#
This example explores different cases where a mesh may not be considered valid as defined
by the validate_mesh() method.
from __future__ import annotations
import pyvista as pv
from pyvista.examples import plot_cell
Non-convex cells#
Many VTK algorithms assume that cells are convex. This can result in incorrect outputs
and may also affect rendering. For example, let’s create PolyData
with a concave QUAD cell.
Use validate_mesh() to show that the cell is not
convex.
report = quad.validate_mesh()
print(report.is_valid)
False
print(report.invalid_fields)
('non_convex',)
If we plot the cell, we can see that the concave cell is incorrectly rendered as though it’s convex even though it is not.
plot_cell(quad, 'xy')
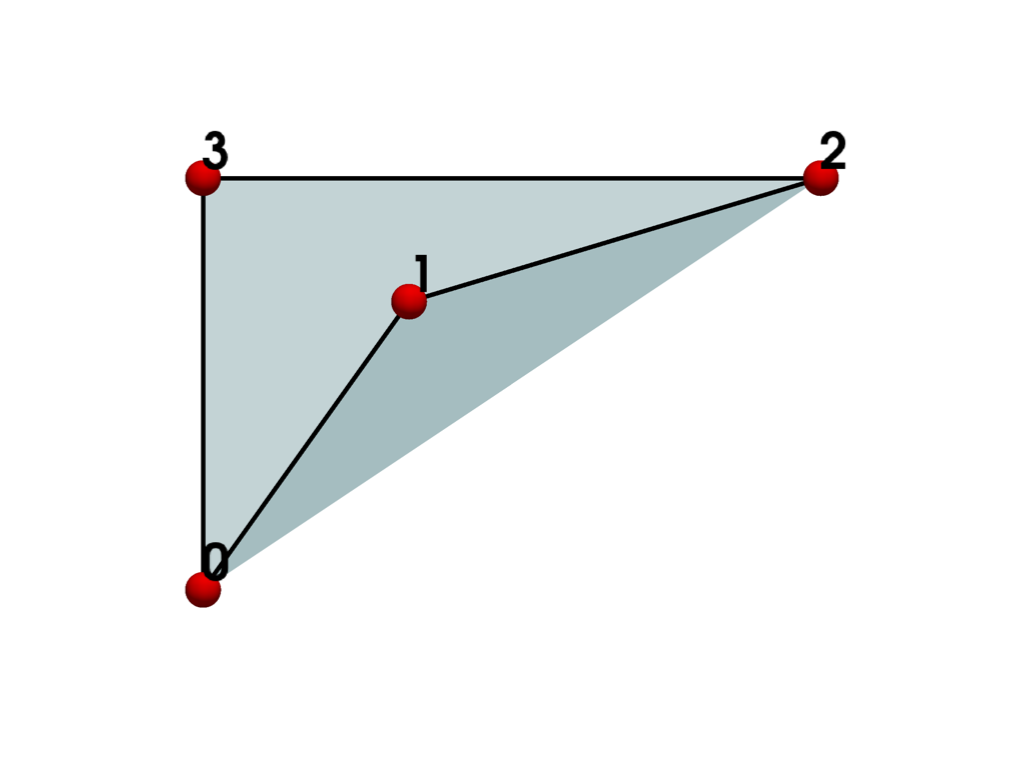
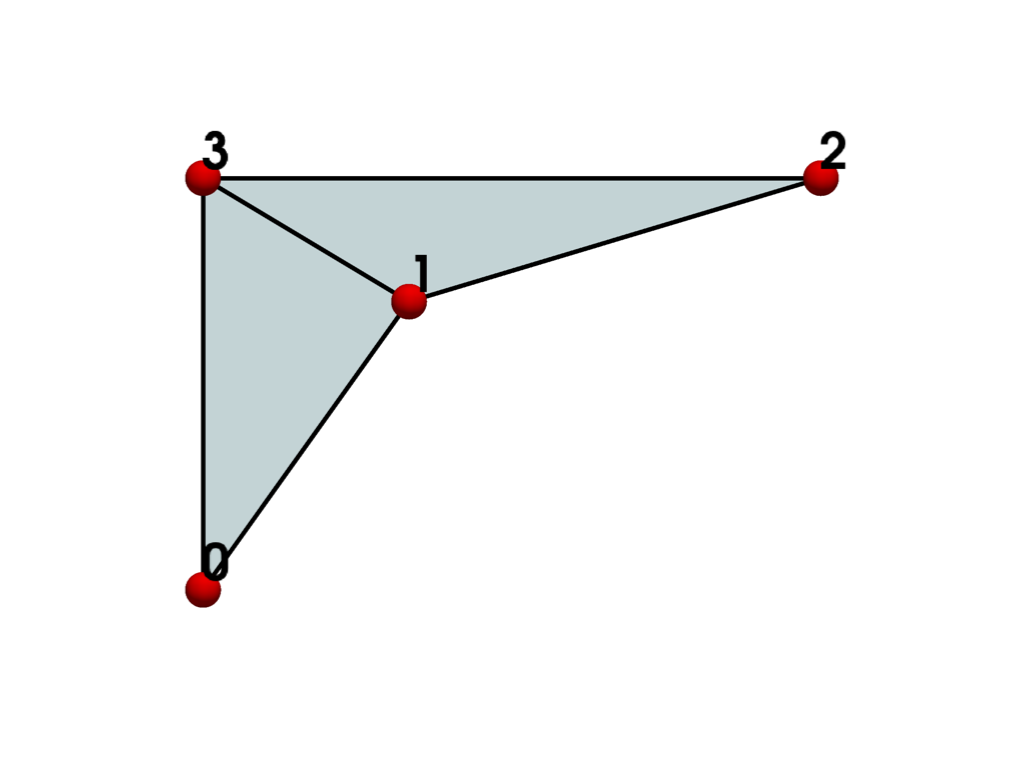
To address the convexity problem, we can triangulate()
the mesh. The mesh is now valid and renders correctly.
triangles = quad.triangulate()
report = triangles.validate_mesh()
print(report.is_valid)
True
plot_cell(triangles, 'xy')
Cells with inverted faces#
Cells with inverted faces can result in incorrect geometric computations such as
cell volume or centroid. To demonstrate this, we first create a valid
POLYHEDRON cell similar to the
Polyhedron() example cell.
points = [[0, 0, 0], [1, 0, 0], [0.5, 0.5, 0], [0, 0, 1]]
cells = [4, 3, 0, 2, 1, 3, 0, 1, 3, 3, 0, 3, 2, 3, 1, 2, 3]
cells = [len(cells), *cells.copy()]
polyhedron = pv.UnstructuredGrid(cells, [pv.CellType.POLYHEDRON], points)
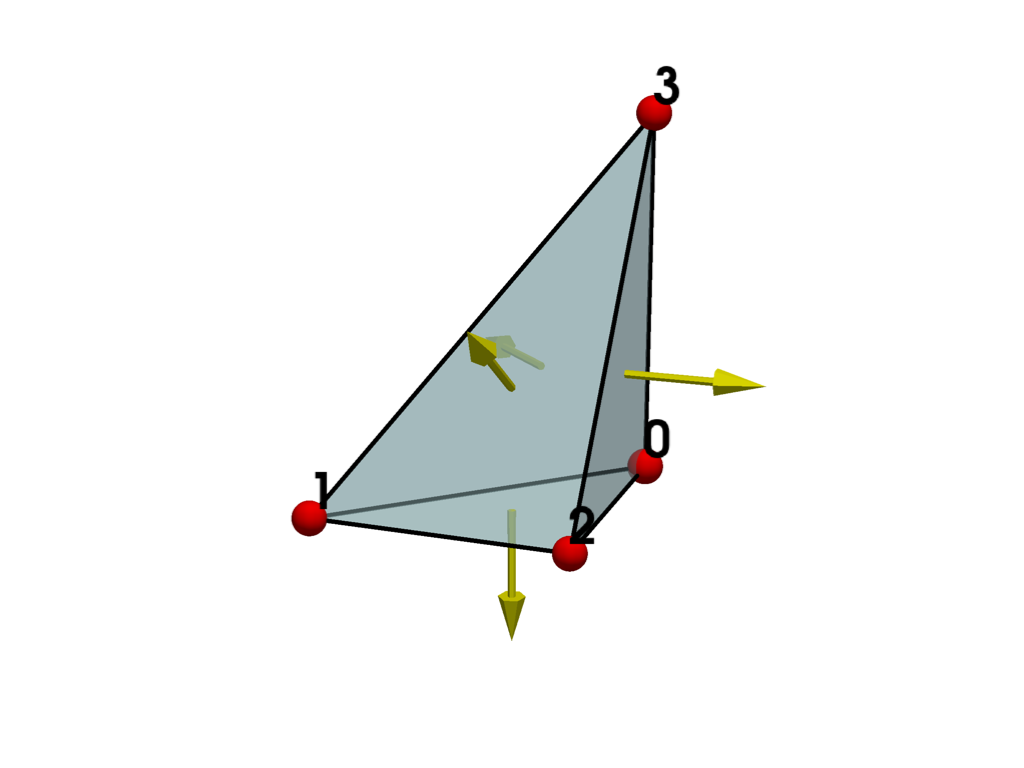
Plot the cell and show its normals. Since all points have counter-clockwise traversal, the normals all point outward and the cell is valid.
report = polyhedron.validate_mesh()
print(report.is_valid)
True
plot_cell(polyhedron, show_normals=True)
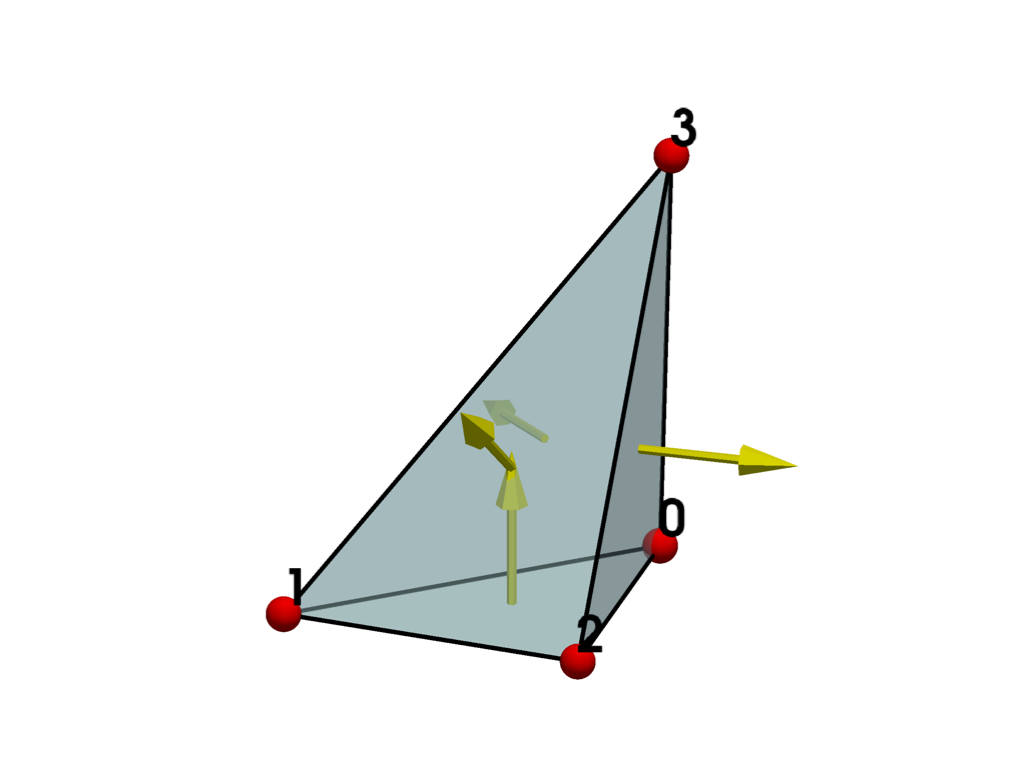
Now swap two points in the polyhedron’s connectivity to generate an otherwise identical polyhedron with a single incorrectly oriented face.
The cell is now invalid, and the bottom face is incorrectly oriented with its normal pointing inward.
report = invalid_polyhedron.validate_mesh()
print(report.is_valid)
False
print(report.invalid_fields)
('inverted_faces',)
plot_cell(invalid_polyhedron, show_normals=True)
Now let’s compare the centroid of the valid and invalid cells using
cell_centers(). The computed centroids differ,
demonstrating the need to have valid cells when using filters that depend on geometric
properties.
valid_centroid = polyhedron.cell_centers().points[0].tolist()
print(valid_centroid)
[0.37499999999999994, 0.12499999999999997, 0.24999999999999994]
invalid_centroid = invalid_polyhedron.cell_centers().points[0].tolist()
print(invalid_centroid)
[0.28125000000000006, 0.09375, 0.43749999999999994]
Self-intersecting cells#
Most cell types have a defined point order which must
be respected. For example, let’s try to create a HEXAHEDRON
cell with eight points:
points = [
[0.0, 0.0, 0.0],
[1.0, 0.0, 0.0],
[0.0, 1.0, 0.0],
[0.0, 0.0, 1.0],
[1.0, 1.0, 0.0],
[1.0, 0.0, 1.0],
[0.0, 1.0, 1.0],
[1.0, 1.0, 1.0],
]
cells = [8, 0, 1, 2, 3, 4, 5, 6, 7]
celltype = [pv.CellType.HEXAHEDRON]
hexahedron = pv.UnstructuredGrid(cells, celltype, points)
At a quick glance, the cell may appear to be valid, but it is not, since the point ordering is incorrect.
report = hexahedron.validate_mesh()
print(report.is_valid)
False
plot_cell(hexahedron)
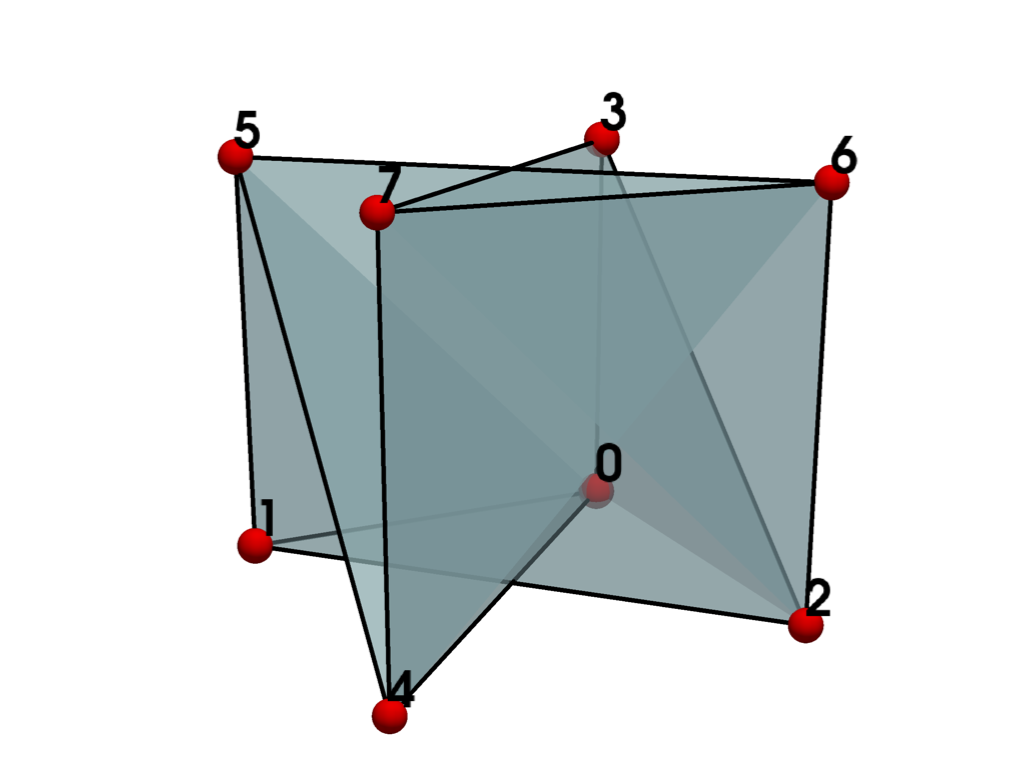
Let’s review the invalid fields reported.
print(report.invalid_fields)
('intersecting_edges', 'inverted_faces', 'non_planar_faces', 'zero_size')
Note that the invalid fields may differ across VTK versions or even operating systems. If a cell is invalid, there are sometimes multiple inter-related issues that may be reported.
From the plot above, we can visually confirm these issues since some faces appear to
intersect, and others appear to be “folded” and hence there are non-planar. To
investigate the 'inverted_faces' problem further, let’s plot the cell again with
normals.
plot_cell(hexahedron, show_normals=True)
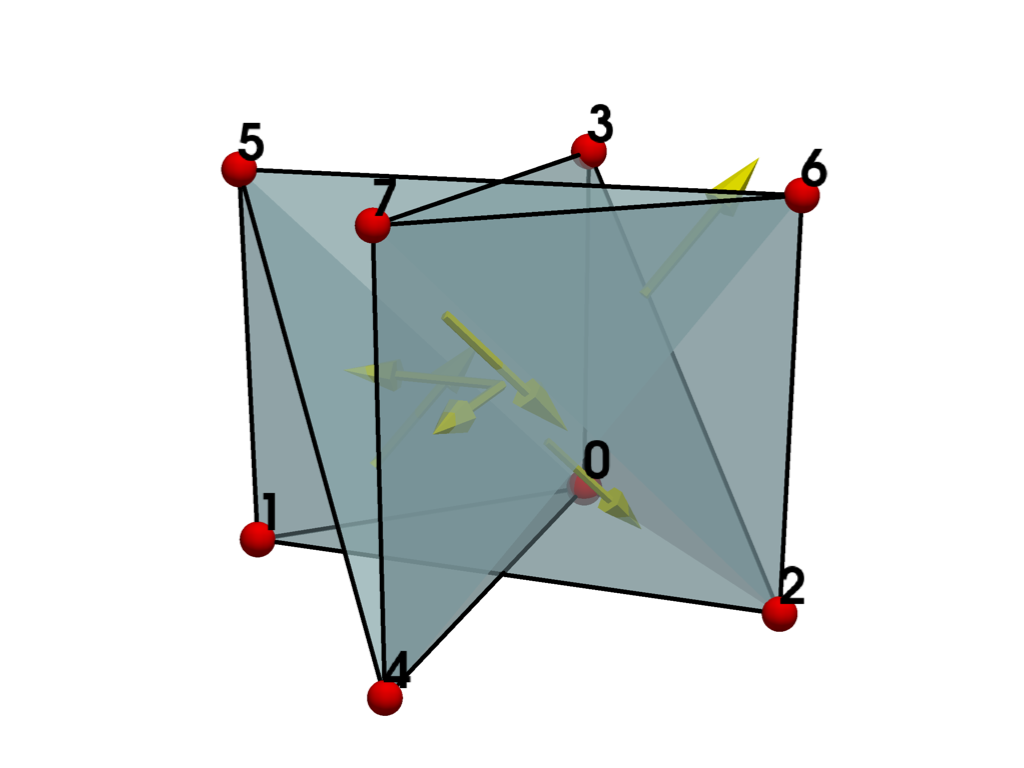
Since some of the normals are pointing inward, this confirms that there are inverted faces. To make the cell valid, we need to re-order the cell connectivity based on the required ordering stated in the documentation for vtkHexahedron.
cells = [8, 0, 1, 4, 2, 3, 5, 7, 6] # instead of [8, 0, 1, 2, 3, 4, 5, 6, 7]
celltype = [pv.CellType.HEXAHEDRON]
hexahedron = pv.UnstructuredGrid(cells, celltype, points)
report = hexahedron.validate_mesh()
print(report.is_valid)
True
plot_cell(hexahedron)
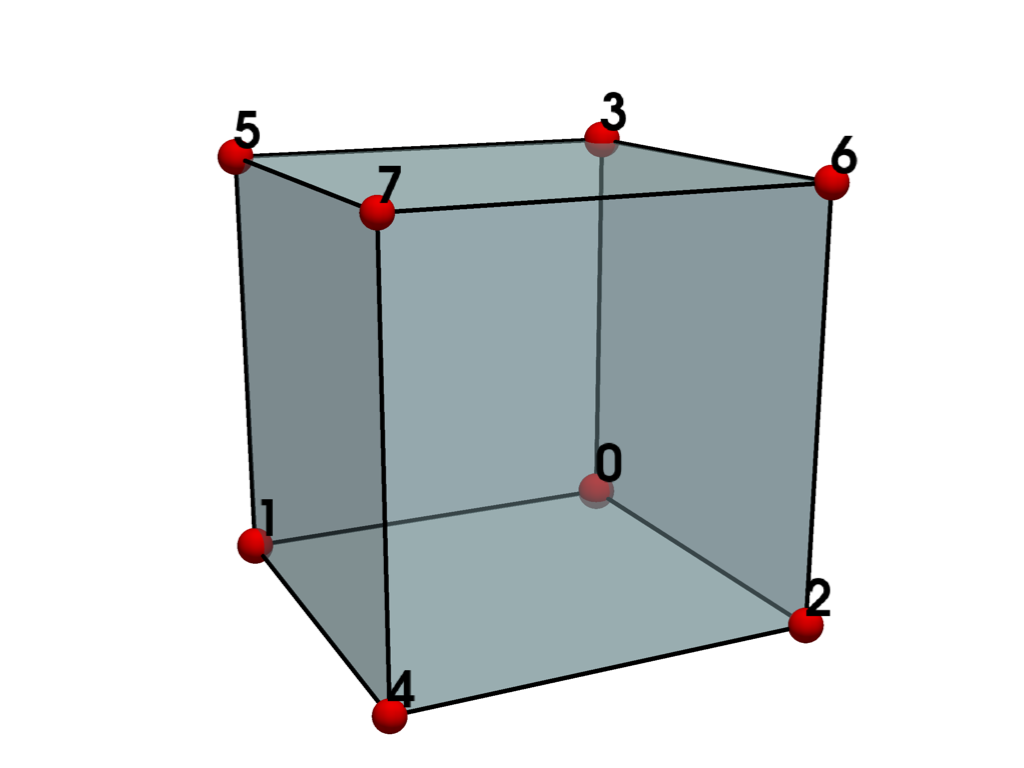
Meshes with unused points#
Unused points are points not associated with any cells. These points are not processed
consistently by filters and are often ignored or removed. To demonstrate this, create an
UnstructuredGrid with a single unused point.
grid = pv.UnstructuredGrid()
grid.points = [[0.0, 0.0, 0.0]]
print(grid.n_points)
1
print(grid.n_cells)
0
This mesh is not considered valid.
report = grid.validate_mesh()
print(report.is_valid)
False
print(report.invalid_fields)
('unused_points',)
Use extract_surface() on the grid and observe that the
unused point is removed.
poly = grid.extract_surface(algorithm=None)
print(poly.n_points)
0
print(poly.n_cells)
0
To remedy this, it is recommended to always associate individual points with a
VERTEX cell. E.g.:
points = [[0.0, 0.0, 0.0]]
cells = [1, 0]
celltypes = [pv.CellType.VERTEX]
grid = pv.UnstructuredGrid(cells, celltypes, points)
print(grid.n_points)
1
print(grid.n_cells)
1
This time, the point is properly processed by the filter and is retained.
poly = grid.extract_surface(algorithm=None)
print(poly.n_points)
1
print(poly.n_cells)
1
This mesh is also now considered valid.
report = grid.validate_mesh()
print(report.is_valid)
True
Total running time of the script: (0 minutes 0.935 seconds)