pyvista.ImageDataFilters.resample#
- ImageDataFilters.resample(
- sample_rate: float | VectorLike[float] | None = None,
- interpolation: _InterpolationOptions = 'nearest',
- *,
- border_mode: Literal['clamp', 'wrap', 'mirror'] = 'clamp',
- reference_image: ImageData | None = None,
- dimensions: VectorLike[int] | None = None,
- anti_aliasing: bool = False,
- extend_border: bool | None = None,
- scalars: str | None = None,
- preference: Literal['point', 'cell'] = 'point',
- inplace: bool = False,
- progress_bar: bool = False,
Resample the image to modify its dimensions and spacing.
The resampling can be controlled in several ways:
Specify the output geometry using a
reference_image.Specify the
dimensionsexplicitly.Specify the
sample_rateexplicitly.
Use
reference_imagefor full control of the resampled geometry. For all other options, the geometry is implicitly defined such that the resampled image fits the bounds of the input.This filter may be used to resample either point or cell data. For point data, this filter assumes the data is from discrete samples in space which represent pixels or voxels; the resampled bounds are therefore extended by 1/2 voxel spacing by default though this may be disabled.
Note
Singleton dimensions are not resampled by this filter, e.g. 2D images will remain 2D.
Added in version 0.45.
- Parameters:
- sample_rate
float|VectorLike[float],optional Sampling rate(s) to use. Can be a single value or vector of three values for each axis. Values greater than
1.0will up-sample the axis and values less than1.0will down-sample it. Values must be greater than0.- interpolation‘nearest’, ‘linear’, ‘cubic’, ‘lanczos’, ‘hamming’, ‘blackman’, ‘bspline’
Interpolation mode to use.
'nearest'(default) duplicates (if upsampling) or removes (if downsampling) values but does not modify them.'linear'and'cubic'use linear and cubic interpolation, respectively.'lanczos','hamming', and'blackman'use a windowed sinc filter and may be used to preserve sharp details and/or reduce image artifacts.'bspline'uses an n-degree basis spline to smoothly interpolate across points. The default degree is3, but can range from0to9. Append the desired degree to the string to set it, e.g.'bspline5'for a 5th-degree B-spline.
Added in version 0.47: Added
'bspline'interpolation.Note
use
'nearest'for pixel art or categorical data such as segmentation masksuse
'linear'for speed-critical tasksuse
'cubic'for upscaling or general-purpose resamplinguse
'lanczos'for high-detail downsampling (at the cost of some ringing)use
'blackman'for minimizing ringing artifacts (at the cost of some detail)use
'hamming'for a balance between detail-preservation and reducing ringing
- border_mode‘clamp’ | ‘wrap’ | ‘mirror’, default: ‘clamp’
Controls the interpolation at the image’s borders.
'clamp'- values outside the image are clamped to the nearest edge.'wrap'- values outside the image are wrapped periodically along the axis.'mirror'- values outside the image are mirrored at the boundary.
Added in version 0.47.
- reference_image
ImageData,optional Reference image to use. If specified, the input is resampled to match the geometry of the reference. The
dimensions,spacing,origin,offset, anddirection_matrixof the resampled image will all match the reference image.- dimensions
VectorLike[int],optional Set the output
dimensionsof the resampled image.Note
Dimensions is the number of points along each axis. If resampling cell data, each dimension should be one more than the number of desired output cells (since there are
Ncells andN+1points along each axis). See examples.- anti_aliasingbool, default:
False Enable antialiasing. This will blur the image as part of the resampling to reduce image artifacts when down-sampling. Has no effect when up-sampling.
- extend_borderbool,
optional Extend the apparent input border by approximately half the
spacing. If enabled, the bounds of the resampled points will be larger than the input image bounds. Enabling this option also has the effect that the re-sampled spacing will directly correlate with the resampled dimensions, e.g. if the dimensions are doubled the spacing will be halved. See examples.This option is enabled by default when resampling point data. Has no effect when resampling cell data or when a
reference_imageis provided.- scalars
str,optional Name of scalars to resample. Defaults to currently active scalars.
- preference
str, default: ‘point’ When scalars is specified, this is the preferred array type to search for in the dataset. Must be either
'point'or'cell'.- inplacebool, default:
False If
True, resample the image inplace. By default, a newImageDatainstance is returned.- progress_barbool, default:
False Display a progress bar to indicate progress.
- sample_rate
- Returns:
ImageDataResampled image.
See also
cropCrop image to remove points at the image’s boundaries.
sample()Resample array data from one mesh onto another.
interpolate()Interpolate values from one mesh onto another.
- Image Data Representations
Compare images represented as points vs. cells.
Examples
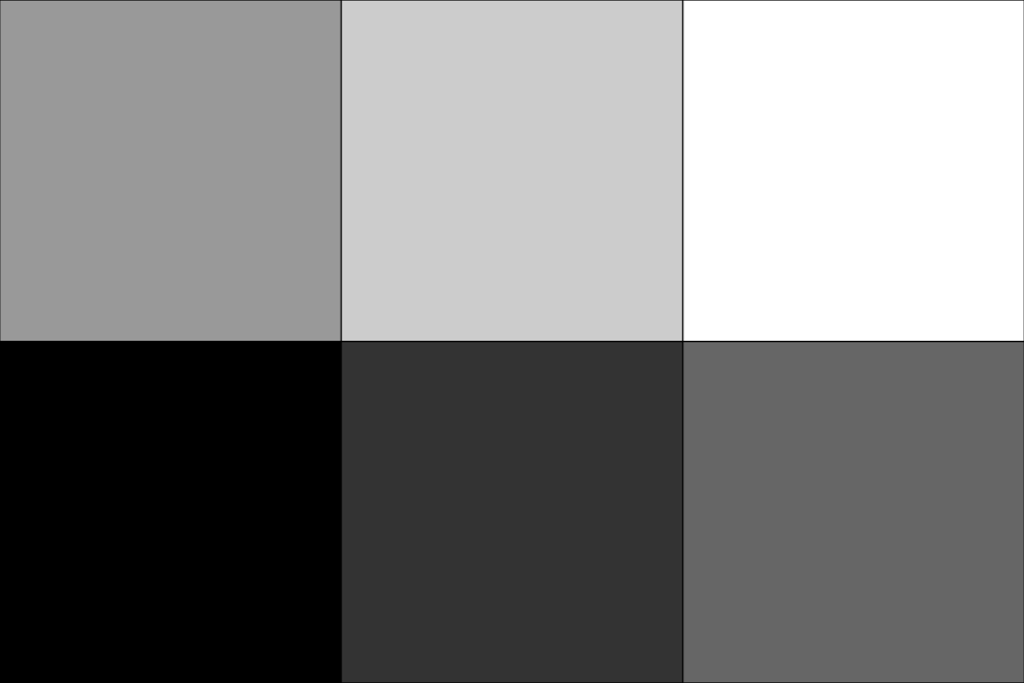
Create a small 2D grayscale image with dimensions
3 x 2for demonstration.>>> import pyvista as pv >>> import numpy as np >>> from pyvista import examples >>> image = pv.ImageData(dimensions=(3, 2, 1)) >>> image.point_data['data'] = np.linspace(0, 255, 6, dtype=np.uint8)
Define a custom plotter to show the image. Although the image data is defined as point data, we use
points_to_cells()to display the image asPIXEL(orVOXEL) cells instead. Grayscale coloring is used and the camera is adjusted to fit the image.>>> def image_plotter(image: pv.ImageData, clim=(0, 255)) -> pv.Plotter: ... pl = pv.Plotter() ... image = image.points_to_cells() ... pl.add_mesh( ... image, ... lighting=False, ... show_edges=True, ... cmap='grey', ... clim=clim, ... show_scalar_bar=False, ... line_width=3, ... ) ... pl.view_xy() ... pl.camera.tight() ... pl.enable_anti_aliasing() ... return pl
Show the image.
>>> plot = image_plotter(image) >>> plot.show()
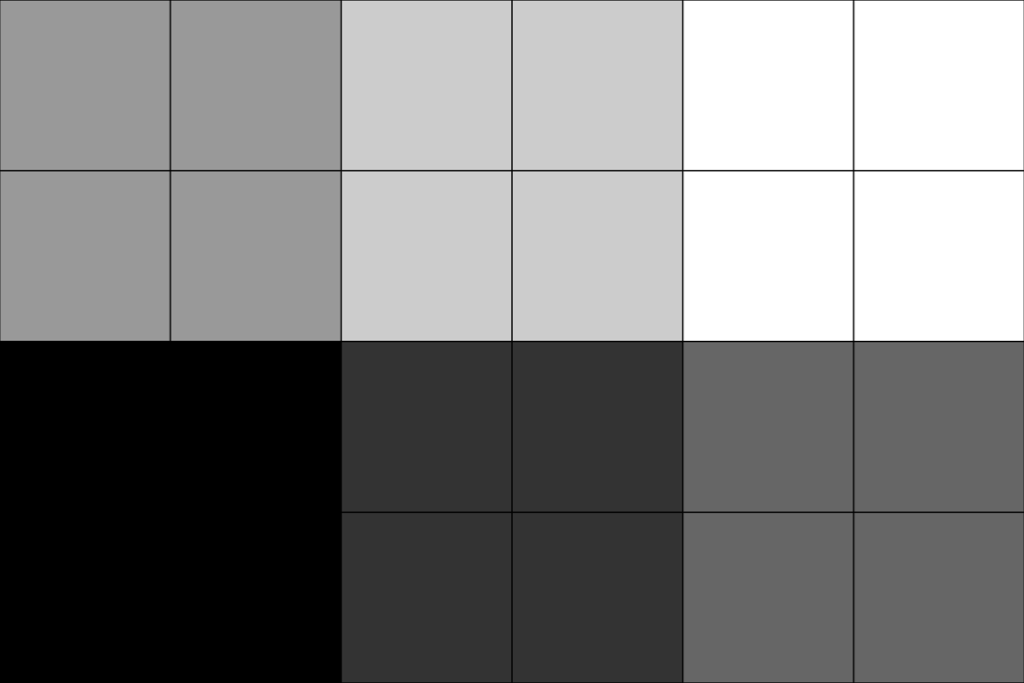
Use
sample_rateto up-sample the image.'nearest'interpolation is used by default.>>> upsampled = image.resample(sample_rate=2.0) >>> plot = image_plotter(upsampled) >>> plot.show()
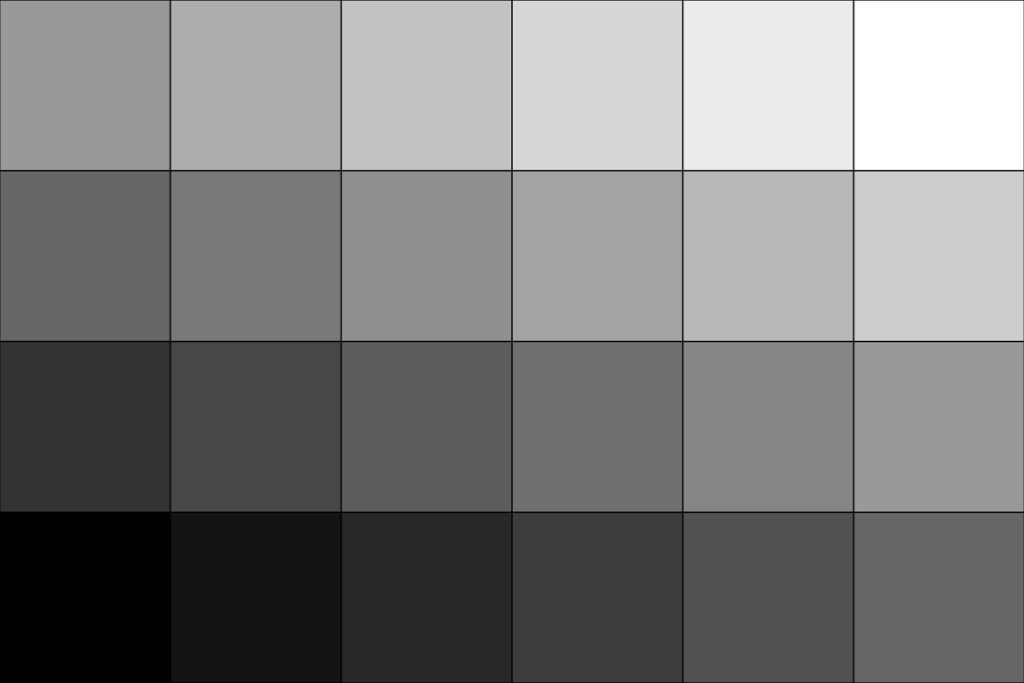
Use
'linear'interpolation. Note that the argument namessample_rateandinterpolationmay be omitted.>>> upsampled = image.resample(2.0, 'linear') >>> plot = image_plotter(upsampled) >>> plot.show()
Use
'cubic'interpolation. Here we also specify the outputdimensionsexplicitly instead of usingsample_rate.>>> upsampled = image.resample(dimensions=(6, 4, 1), interpolation='cubic') >>> plot = image_plotter(upsampled) >>> plot.show()
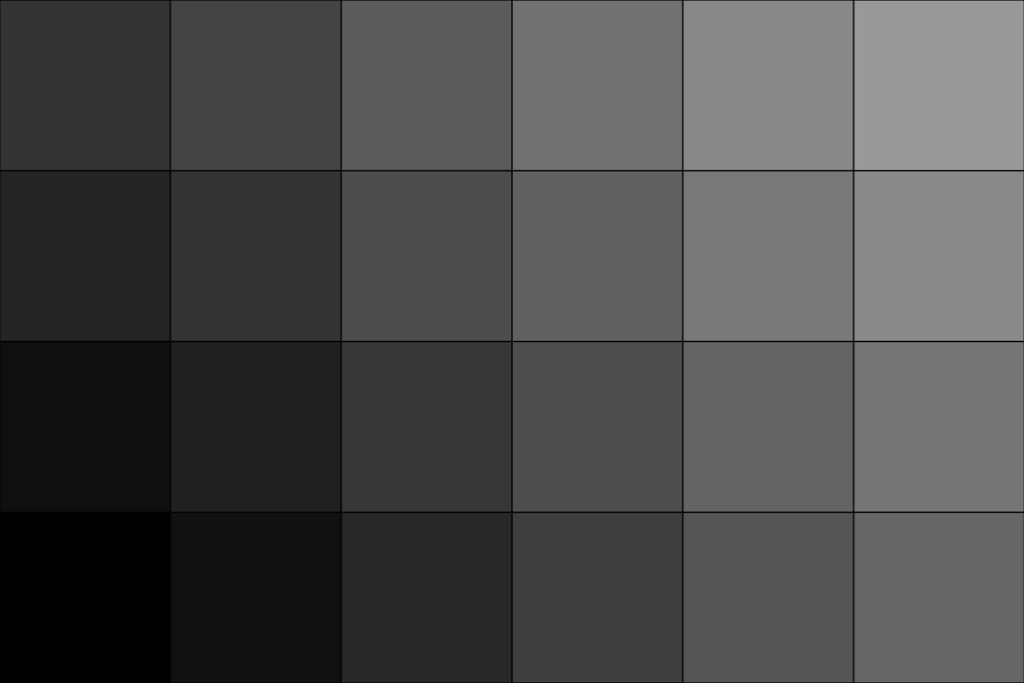
Compare the relative physical size of the image before and after resampling.
>>> image ImageData (...) N Cells: 2 N Points: 6 X Bounds: 0.000e+00, 2.000e+00 Y Bounds: 0.000e+00, 1.000e+00 Z Bounds: 0.000e+00, 0.000e+00 Dimensions: 3, 2, 1 Spacing: 1.000e+00, 1.000e+00, 1.000e+00 N Arrays: 1
>>> upsampled ImageData (...) N Cells: 15 N Points: 24 X Bounds: -2.500e-01, 2.250e+00 Y Bounds: -2.500e-01, 1.250e+00 Z Bounds: 0.000e+00, 0.000e+00 Dimensions: 6, 4, 1 Spacing: 5.000e-01, 5.000e-01, 1.000e+00 N Arrays: 1
Note that the upsampled
dimensionsare doubled and thespacingis halved (as expected). Also note, however, that the physical bounds of the input differ from the output. The upsampledoriginalso differs:>>> image.origin (0.0, 0.0, 0.0) >>> upsampled.origin (-0.25, -0.25, 0.0)
This is because the resampling is done with
extend_borderenabled by default which adds a half cell-width border to the image and adjusts the origin and spacing such that the bounds match when the image is represented as cells.Apply
points_to_cells()to the input and resampled images and show that the bounds match.>>> image_as_cells = image.points_to_cells() >>> image_as_cells.bounds BoundsTuple(x_min = -0.5, x_max = 2.5, y_min = -0.5, y_max = 1.5, z_min = 0.0, z_max = 0.0)
>>> upsampled_as_cells = upsampled.points_to_cells() >>> upsampled_as_cells.bounds BoundsTuple(x_min = -0.5, x_max = 2.5, y_min = -0.5, y_max = 1.5, z_min = 0.0, z_max = 0.0)
Plot the two images together as wireframe to visualize them. The original is in red, and the resampled image is in black.
>>> pl = pv.Plotter() >>> _ = pl.add_mesh( ... image_as_cells, style='wireframe', color='red', line_width=10 ... ) >>> _ = pl.add_mesh( ... upsampled_as_cells, style='wireframe', color='black', line_width=2 ... ) >>> pl.view_xy() >>> pl.camera.tight() >>> pl.show()
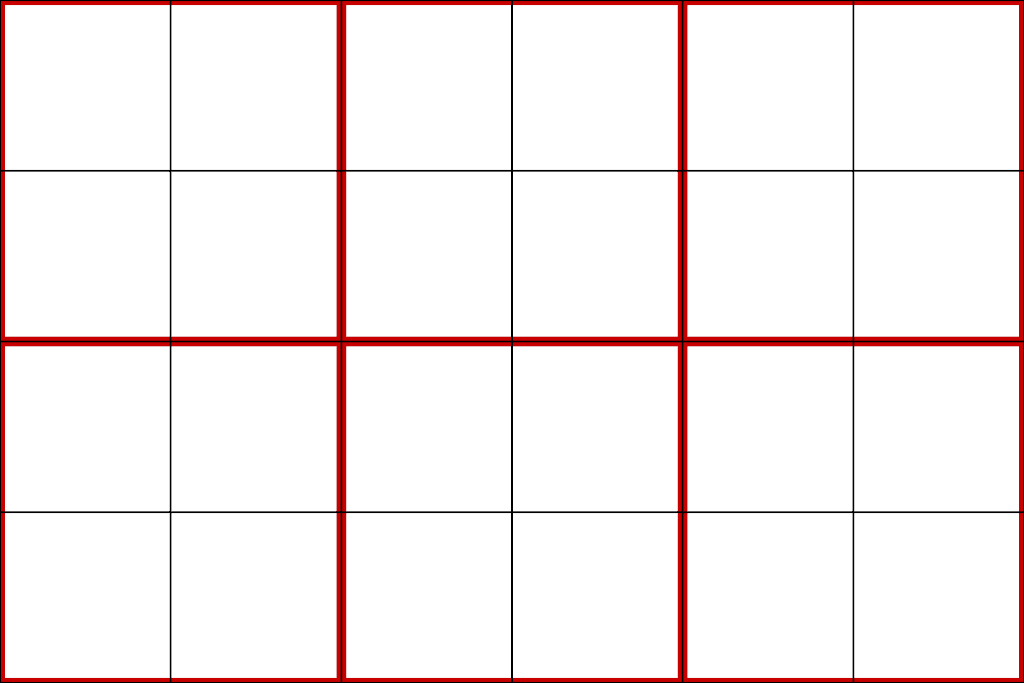
Disable
extend_borderto force the input and output bounds of the points to be the same instead.>>> upsampled = image.resample(sample_rate=2, extend_border=False)
Compare the two images again.
>>> image ImageData (...) N Cells: 2 N Points: 6 X Bounds: 0.000e+00, 2.000e+00 Y Bounds: 0.000e+00, 1.000e+00 Z Bounds: 0.000e+00, 0.000e+00 Dimensions: 3, 2, 1 Spacing: 1.000e+00, 1.000e+00, 1.000e+00 N Arrays: 1
>>> upsampled ImageData (...) N Cells: 15 N Points: 24 X Bounds: 0.000e+00, 2.000e+00 Y Bounds: 0.000e+00, 1.000e+00 Z Bounds: 0.000e+00, 0.000e+00 Dimensions: 6, 4, 1 Spacing: 4.000e-01, 3.333e-01, 1.000e+00 N Arrays: 1
This time the input and output bounds match without any further processing. Like before, the dimensions have doubled; unlike before, however, the spacing is not halved, but is instead smaller than half which is necessary to ensure the bounds remain the same. Also unlike before, the origin is unaffected:
>>> image.origin (0.0, 0.0, 0.0) >>> upsampled.origin (0.0, 0.0, 0.0)
All the above examples are with 2D images with point data. However, the filter also works with 3D volumes and will also work with cell data.
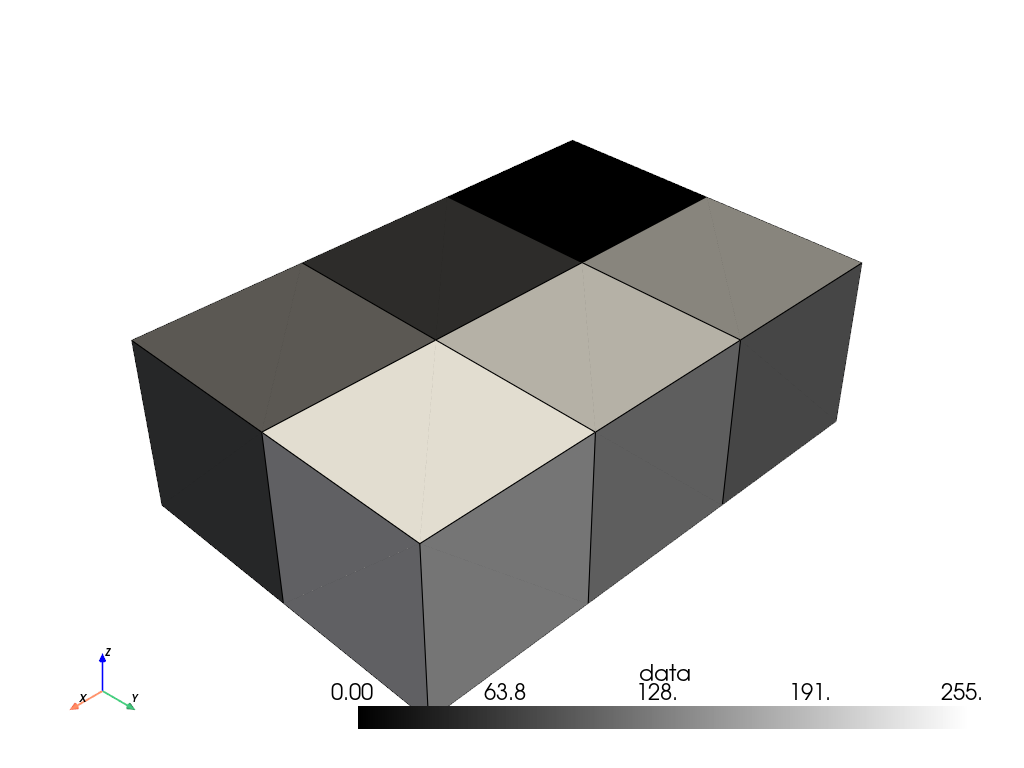
Convert the 2D image with point data into a 3D volume with cell data and plot it for context.
>>> volume = image.points_to_cells(dimensionality='3D') >>> volume.plot(show_edges=True, cmap='grey')
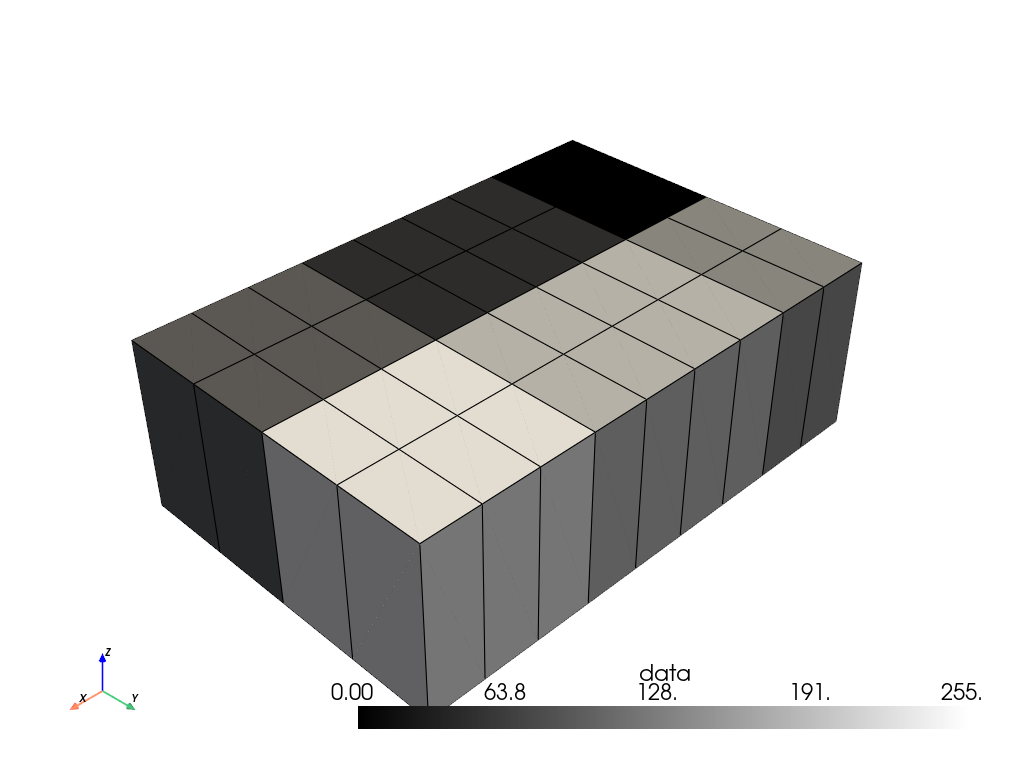
Up-sample the volume. Set the sampling rate for each axis separately.
>>> resampled = volume.resample(sample_rate=(3.0, 2.0, 1.0)) >>> resampled.plot(show_edges=True, cmap='grey')
Alternatively, we could have set the dimensions explicitly. Since we want
9 x 4 x 1cells along the x-y-z axes (respectively), we set the dimensions to(10, 5, 2), i.e. one more than the desired number of cells.>>> resampled = volume.resample(dimensions=(10, 5, 2)) >>> resampled.plot(show_edges=True, cmap='grey')
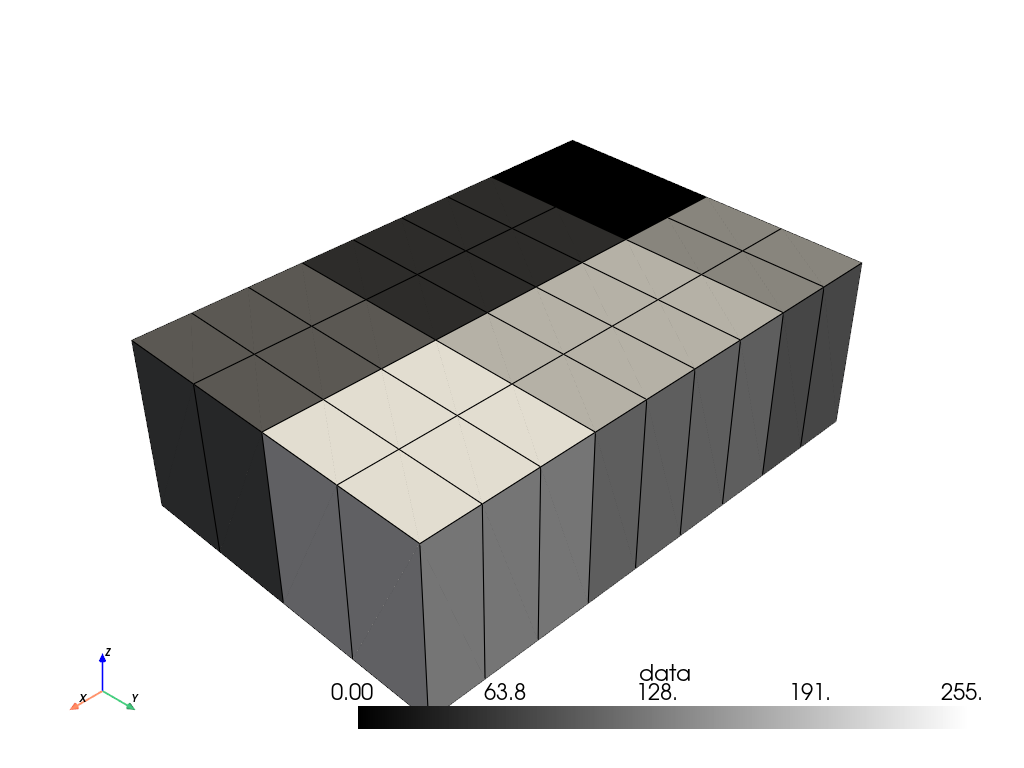
Compare the bounds before and after resampling. Unlike with point data, the bounds are not (and cannot be) extended.
>>> volume.bounds BoundsTuple(x_min = -0.5, x_max = 2.5, y_min = -0.5, y_max = 1.5, z_min = -0.5, z_max = 0.5) >>> resampled.bounds BoundsTuple(x_min = -0.5, x_max = 2.5, y_min = -0.5, y_max = 1.5, z_min = -0.5, z_max = 0.5)
Use a reference image to control the resampling instead. Here we load two images with different dimensions:
download_bird()anddownload_gourds().>>> bird = examples.download_bird() >>> bird.dimensions (458, 342, 1)
>>> gourds = examples.download_gourds() >>> gourds.dimensions (640, 480, 1)
Use
reference_imageto resample the bird to match the gourds geometry or vice-versa.>>> bird_resampled = bird.resample(reference_image=gourds) >>> bird_resampled.dimensions (640, 480, 1)
>>> gourds_resampled = gourds.resample(reference_image=bird) >>> gourds_resampled.dimensions (458, 342, 1)
Downsample the gourds image to 1/10th its original resolution using
'lanczos'interpolation.>>> downsampled = gourds.resample(1 / 8, 'lanczos') >>> downsampled.dimensions (80, 60, 1)
Compare the downsampled image to the original and zoom in to show detail.
>>> def compare_images_plotter(image1, image2): ... pl = pv.Plotter(shape=(1, 2)) ... _ = pl.add_mesh(image1, rgba=True, show_edges=False, lighting=False) ... pl.subplot(0, 1) ... _ = pl.add_mesh(image2, rgba=True, show_edges=False, lighting=False) ... pl.link_views() ... pl.view_xy() ... pl.camera.zoom(3.0) ... return pl
>>> pl = compare_images_plotter(gourds, downsampled) >>> pl.show()

Note that downsampling can create image artifacts caused by aliasing. Enable anti-aliasing to smooth the image before resampling.
>>> downsampled2 = gourds.resample(1 / 8, 'lanczos', anti_aliasing=True)
Compare down-sampling with aliasing (left) to without aliasing (right).
>>> pl = compare_images_plotter(downsampled, downsampled2) >>> pl.show()

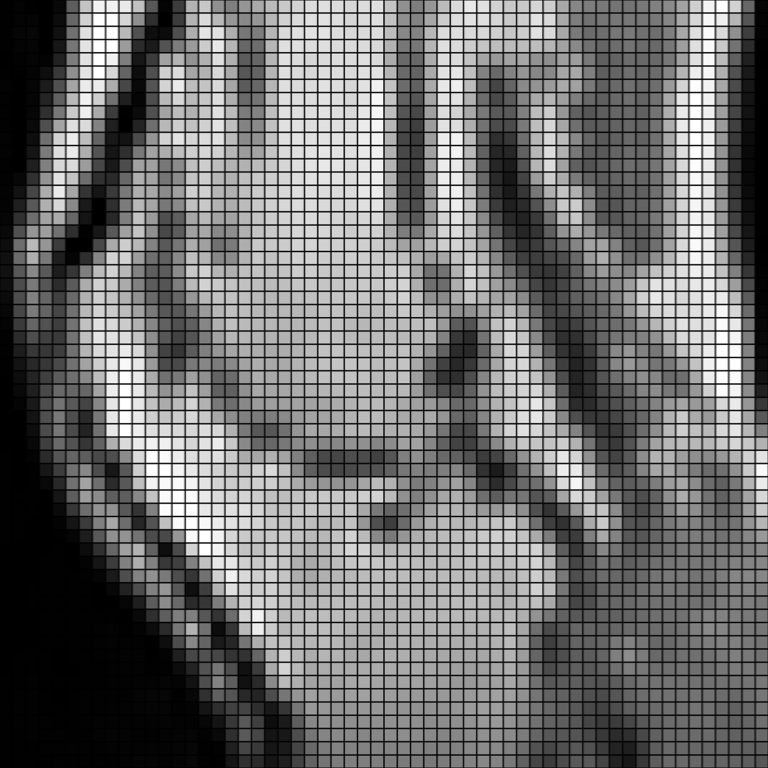
Load an MRI of a knee and downsample it.
>>> knee = pv.examples.download_knee().resample( ... 0.1, 'linear', anti_aliasing=True ... )
Crop and plot it.
>>> knee = knee.crop(normalized_bounds=[0.2, 0.8, 0.2, 0.8, 0.0, 1.0]) >>> vmin = knee.active_scalars.min() >>> vmax = knee.active_scalars.max() >>> pl = image_plotter(knee, clim=[vmin, vmax]) >>> pl.show()

Upsample it with B-spline interpolation. The interpolation is very smooth.
>>> upsampled = knee.resample(2.0, 'bspline', border_mode='clamp') >>> pl = image_plotter(upsampled, clim=[vmin, vmax]) >>> pl.show()
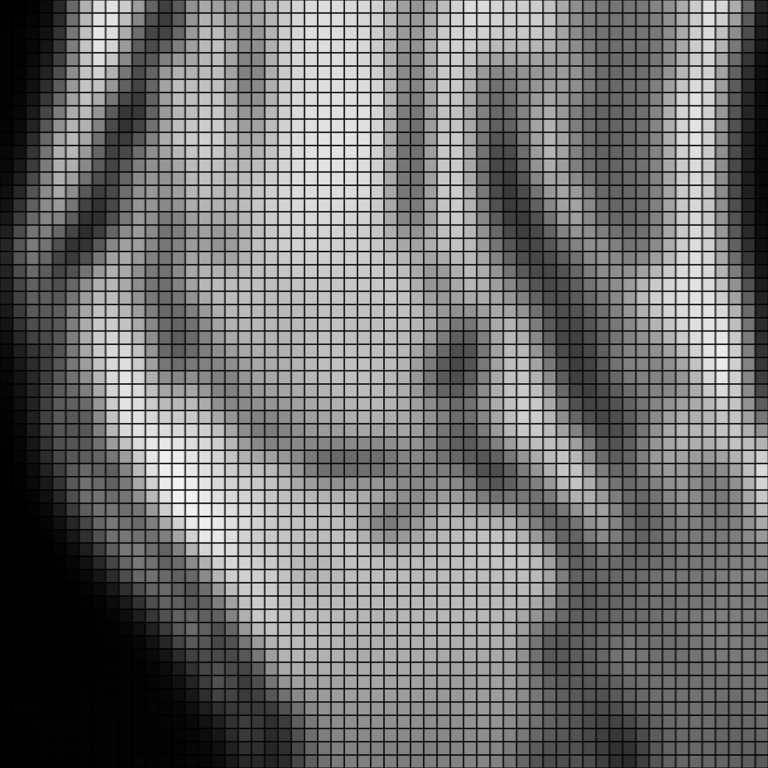
Use the
'wrap'border mode. Note how points at the border are brighter than previously, since the bright pixels from the opposite edge are now included in the interpolation.>>> upsampled = knee.resample(2.0, 'bspline', border_mode='wrap') >>> pl = image_plotter(upsampled, clim=[vmin, vmax]) >>> pl.show()
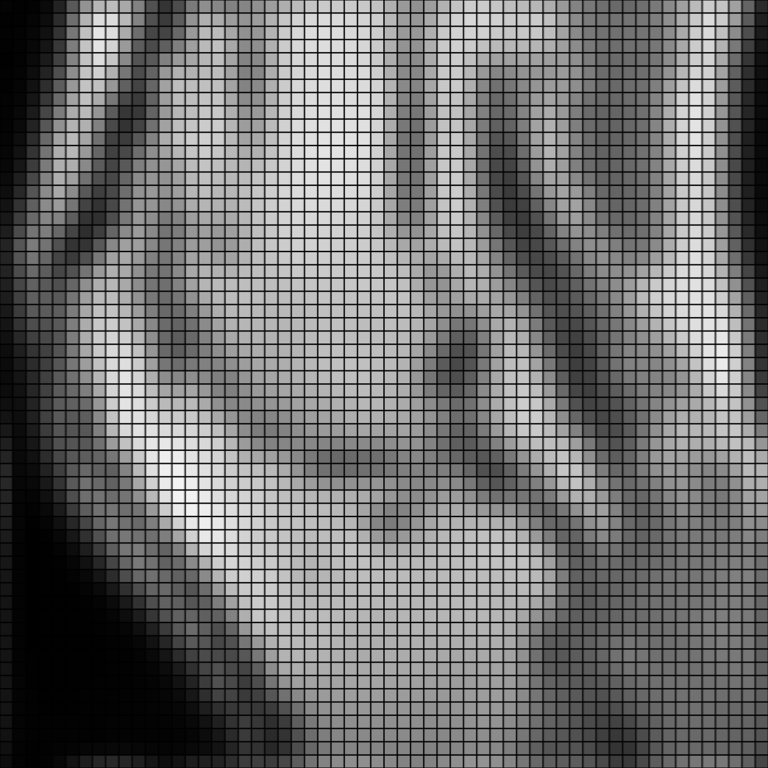
Compare B-spline interpolation to
'hamming'.>>> upsampled = knee.resample(2.0, 'hamming') >>> pl = image_plotter(upsampled, clim=[vmin, vmax]) >>> pl.show()